A brief example of neural data analysis using MATLAB
Edgar Bermudez, PhD
March 2013
Neural data analysis gives a better idea about the response or information processing/computation in a group of neurons.
Contents
Electrophysiology experiments
The recording system keeps track of
- electrical signals collected from electrodes/silicon probes and also
- stimulation information.

Silicon probes have recording sites (called tetrodes) with different configurations. Each probe has an array of shanks with recording sites. For the recordings in this experiment, one probe was inserted in S1 and another in mPFC.
Experiment description
The experiment consisted in recording neuronal data in anesthetized rats receving tactile stimulation under two conditions:
- only urethane and
- urethane + amphetamine

Neural data was recorded during periods of spontaneous (no stimulation) and evoked activity (where tactile stimulation was given) for both conditions.
Example of neuronal activity

There are different ways to represent neuronal activity, for this example, we choose to store spike information in a matrix with zeros and ones.
Important data structures in a electrophysiology recording
- SpkCnt : neurons x time
- stim : times of tactile stimulation
- SpkEle : neurons - tetrode
- stim : time - event type
For this simple example we are going to analyze only a reduced sample of spike data (due to memory restrictions in the lab), however in a normal analysis we can combine LFP and spike data in recordings which can be very lengthy.
Plot experiment setup

1: SP ure, 2: stim ure, 3: SP ure, 5:SP Amph, 6:stim Amph, 7:SP Amph
If you are interested in looking at the same analysis for the whole recordings, check http://people.uleth.ca/~edgar.bermudez/matlab_class/html/matlabclass_analysis.html
Stimulus triggered activity
In order to have an idea about how neurons in the different areas respond to stimulation, we can display the stimulus triggered response (PSTH).
Load the data
datadir = 'C:\Users\edgar.bermudez\Documents\'; load([datadir 'LK10_sm' ]);
Neurons for S1 and PFC
pfc_nns = find( SpkEle<=8); % pfc neurons s1_nns = find( SpkEle > 8); % s1 neurons nr_pfc_nns = length( pfc_nns ); % number of pfc neurons
Extracting useful information In order to target our analysis, we divide and label the data to have a better control. For example, we need to know:
- the times of both conditions
- when stimulation was given
- how many times stimulation was given and take data around it
Some useful info
% Firing rate for S1 neurons during each stage fr1 = mean(SpkCnt1(:,s1_nns)); fr2 = mean(SpkCnt2(:,s1_nns)); fr3 = mean(SpkCnt3(:,s1_nns)); % Firing rate for PFC neurons during each stage fr4 = mean(SpkCnt1(:,pfc_nns)); fr5 = mean(SpkCnt2(:,pfc_nns)); fr6 = mean(SpkCnt3(:,pfc_nns));
Selection of active S1 neurons
select neurons with FR above threshold s1 and PFC
fr_th = 0.002; act_s1nns = find( fr1 > fr_th & fr2 > fr_th & fr3 > fr_th); act_pfcnns = find( fr4 > fr_th & fr5 > fr_th & fr6 > fr_th);
Slicing activity into trial locked time windows
% tactile stim trig activity before drug f_tac1 = find(stim_sm==3); f_tac1( find(diff( f_tac1 ) < 10 | diff( f_tac1 ) > 700)+1) = []; w1=100; w2=750; spk1=zeros(length(f_tac1),w1+w2+1,length(act_s1nns)); spk2=zeros(length(f_tac1),w1+w2+1,length(act_pfcnns)); for tr=2:length(f_tac1)-1 spk1(tr,:,:)=SpkCnt2(f_tac1(tr)-w1:f_tac1(tr)+w2,s1_nns(act_s1nns)); spk2(tr,:,:)=SpkCnt2(f_tac1(tr)-w1:f_tac1(tr)+w2,s1_nns(act_pfcnns)); end % range of the reponse plots in ms range = (-100:750)*3.2;
Plotting the response
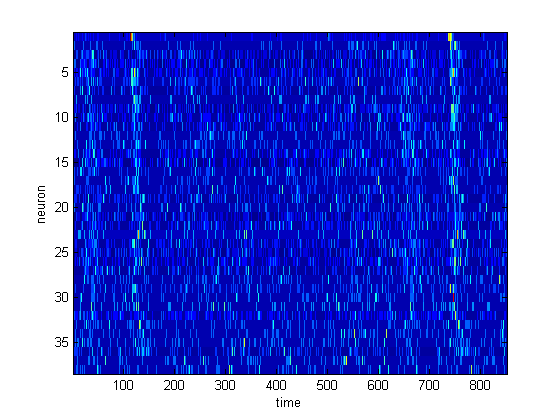
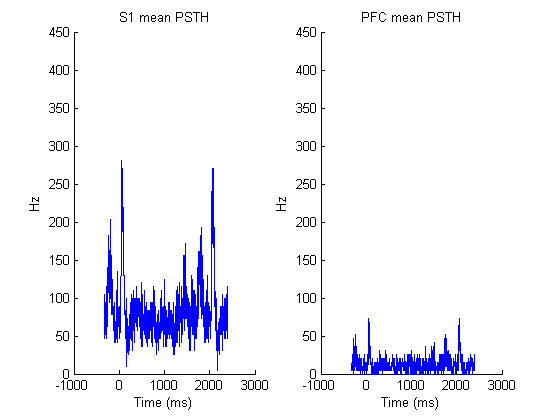
figure(); title(['Neural response in urethane condition']); imagesc(zscore(squeeze(mean(spk1)))'); xlabel('time'); ylabel('neuron'); figure(); subplot(1,2,1); hold on; title(['S1 mean PSTH']); plot(range, 312.5*squeeze(mean(sum(spk1(:,:,:),3)))); ylim([0 450]); xlabel('Time (ms)'); ylabel(' Hz '); subplot(1,2,2); hold on; title(['PFC mean PSTH']); plot(range, 312.5*squeeze(mean(sum(spk2(:,:,:),3)))); ylim([0 450]); xlabel('Time (ms)'); ylabel(' Hz ');
Template matching
Template matching quantifies replay of temporal sequences of neuronal activity.

Template construction
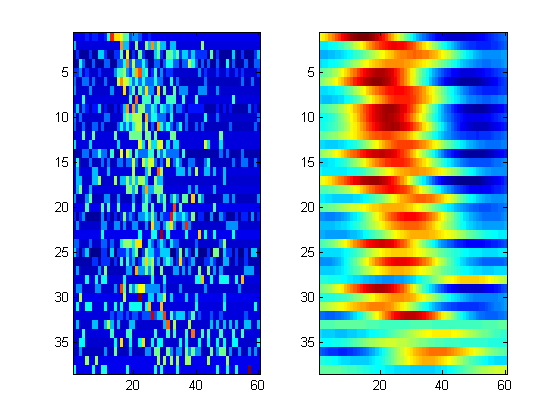
Template is defined as the (summed evoked act - mean) / std average stim triggered activity of the S1 neurons
t_len = 60; t_window = 101:100+t_len; X = squeeze(sum(spk1(:,t_window,:),1))'; zs = zscore(X')'; figure('name','Template'); subplot(1,2,1); imagesc(zs); % In order to have a less specific template, we smooth the activity pattern % for every neuron g_sigma = 20; gs = normpdf(-g_sigma*2:g_sigma*2,0, g_sigma/2 )'; for n=1:size(zs,1) temp = zs( n, :); tmplt(n ,:) = conv2( temp', gs, 'same'); end template = tmplt(:,:); subplot(1,2,2); imagesc(template);
Matching
shift = 1; num_windows = 37000; norm_bef = zeros(1,num_windows); norm_aft = zeros(1,num_windows); % take windows of activity to compare against our template for j=1:num_windows winanalysis = 1+((j-1)*shift) : 1+((j-1)*shift)+t_len-1; % activity before stimulation tgt = SpkCnt1(winanalysis, s1_nns(act_s1nns))'; % we normalize in the same way such windows tgt_z = zscore(tgt')'; norm_aft(j) = dot(template(:), tgt_z(:)); % similarity comparison % activity after stimulation tgt = SpkCnt3(winanalysis, s1_nns(act_s1nns))'; tgt_z = zscore(tgt')'; norm_bef(j) = dot(template(:), tgt_z(:)); % similarity comparison end
Results
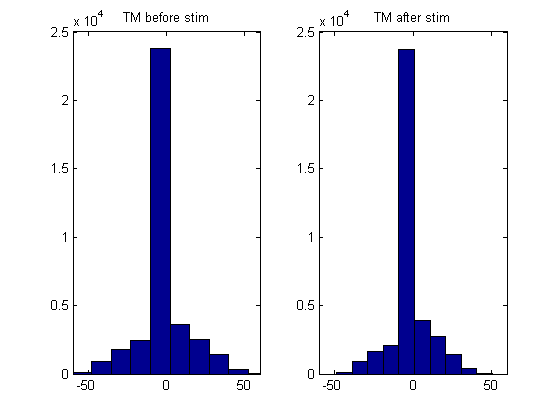
We show the distribution of the TM signal before and after stimulation
figure(); subplot(1,2,1); hist(norm_bef); title('TM before stim'); xlim([-60 60]); subplot(1,2,2); hist(norm_aft); title('TM after stim'); xlim([-60 60]);
References for further reading
- Tatsuno, M., Lipa, P., & McNaughton, B.L. (2006) Methodological considerations on the use of template matching to study long-lasting memory trace replay. The Journal of Neuroscience, 26:10727-10742
- Carr, Margaret F., Jadhav, Shantanu P., Frank, Loren M. (2011) Hippocampal replay in the awake state: a potential substrate for memory consolidation and retrieval. Nature Neuroscience 14, 147–153